-
Notifications
You must be signed in to change notification settings - Fork 28
2.0 refactor #161
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
2.0 refactor #161
Conversation
|
This looks good to me and I am +1 for merging it, however, I'm assuming that this is a WIP, since just the single group measures were refactored, right? |
|
sweet. yeah, still WIP, just wanted to get some feedback/buy-in for this approach. I'll keep going with the multigroup indices |
|
this is ready to go. The main benefit is we adopt the logic of Reardon & Osullivan, that we can derive generalized spatial indices by transforming the input data of aspatial indices through a weights matrix (so, e.g. now we can compute multiscalar profiles for any the original tests are unchanged to show that the (aspatial) calculations are still correct, with one exception: MinMaxS has a very different estimate. I think its correct, but want to flag this one did change aside from the reorganization, there's one important API change, which is that fitted indices have the attribute |
Codecov Report
@@ Coverage Diff @@
## master #161 +/- ##
===========================================
- Coverage 85.75% 65.77% -19.98%
===========================================
Files 65 116 +51
Lines 2779 4322 +1543
===========================================
+ Hits 2383 2843 +460
- Misses 396 1479 +1083
Continue to review full report at Codecov.
|
|
(tests are all passing, there's a CI issue with rtree on windows) |
| statistics : np.array(n,k) | ||
| Local Diversity values for each group and unit | ||
|
|
||
| core_data : a pandas DataFrame |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
The PR looks good to me. But wasn't supposed to return data rather than core_data?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
its ok here because we set the class attribute appropriately (so even though we return the variable core_data above, its contents get set to the class attribute named data)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Cool, gotcha!
|
Awesome, @knaaptime! By the way, when you say "MinMaxS has a very different estimate", you mean the "MinMax" from https://github.com/knaaptime/segregation/blob/2.0/segregation/singlegroup/minmax.py, right? Any clue of why this might be happening? 'Cause it seems like nothing changed in the code/math behind it, right? |
|
right, the math and the function that actually compute the statistic havent changed. I thnk the root is this line in the new version, whereas the old code probably created a Kernel weights object from unprojected data |
Yep, this might it, perhaps the new version of |
|
puts gp at 0.9.0 and problem with |
|
thanks martin |
| c = 1-dist.copy() | ||
|
|
||
| Pxx = (data.xi.values * data.xi.values * c).sum() / (X ** 2) | ||
| Pyy = (data.xi.values * data.yi.values * c).sum() / (X*Y) |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@renanxcortes one substantive change here: I edited this formula because i think the last version had a mistake (ycy/y^2 instead of ycx/yx)
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@knaaptime , the original version is the one that follows Massey and Denton. If we look closer at formulas (19) and (20) which are, respectively, Spatial Proximity and Relative Clustering, they both use the definition of Pxx and Pyy, which is our original implementation.
The definition of the "interaction" represented by "Pxy" is just for illustration.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
ah, snap. you're right
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
No sweat!
Add deprecation layer
add deprecation layer
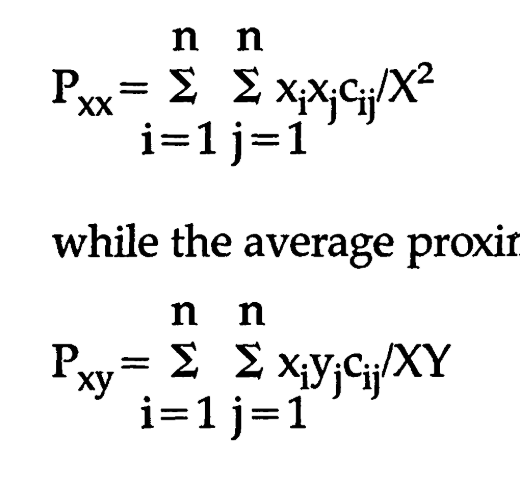

this starts the restructuring discussed in #4. i think it pretty substantially cuts down on repetition and makes the code much easier to navigate. We also get #104 for free